Our
Research
Research at the Rabadan Lab
Our main scientific interests lie in modeling and understanding the dynamics of biological systems through the lens of genomics. We are a very interdisciplinary team of mathematicians, physicists, engineers, biologists, and medical doctors with a common goal of solving pressing medical problems. We are currently focusing our work on:
Cancer. Genomic technologies provide an extraordinary opportunity to identify mutations that contribute to the development of tumors. We are mapping the evolution of cancers and uncovering the mechanisms of response or lack of response to multiple therapies. We work with clinicians and experimentalists all around the world.
Infectious diseases. Evolution is a dynamic process that shapes genomes. Our team at Columbia is developing algorithms to analyze genomic data, with a view to understanding the molecular biology, population genetics, phylogeny, and epidemiology of viruses. We are interested in the emergence of infectious diseases, pandemics and uncovering the mechanisms of adaptation of viruses to humans.
Electronic Health Records. Clinical databases constitute a rich and complex source of raw data. We are using the power of statistics and computers to tease out important clinical patterns in these diverse, important datasets. Combining molecular and clinical data illuminates some of the mechanisms underlying complex diseases.
In particular, we develop mathematical, statistical, and computational approaches, which cover the analysis of high throughput data right through to the altogether more abstract identification of global patterns in evolutionary processes. Learn more about the Rabadan Lab and the three main global questions that we are addressing.
Research Projects
Illuminating the Noncoding Genome in Cancer using Artificial Intelligence.
Understanding the vast noncoding cancer genome requires cutting-edge, high-resolution, and accessible strategies. Artificial intelligence (AI) is revolutionizing cancer research, enabling advanced models to analyze genome regulation. This review examines illustrative examples of noncoding mutations in cancer, focusing on both key regulatory elements and risk-associated variants that remain poorly understood, and compares key AI models developed over the last decade for identifying functional noncoding variants, predicting gene expression impacts, and uncovering cancer-associated mutations. The discussion of the goals, data requirements, features, and outcomes of the models offers practical insights to help cancer researchers integrate these technologies into their work, regardless of computational expertise.
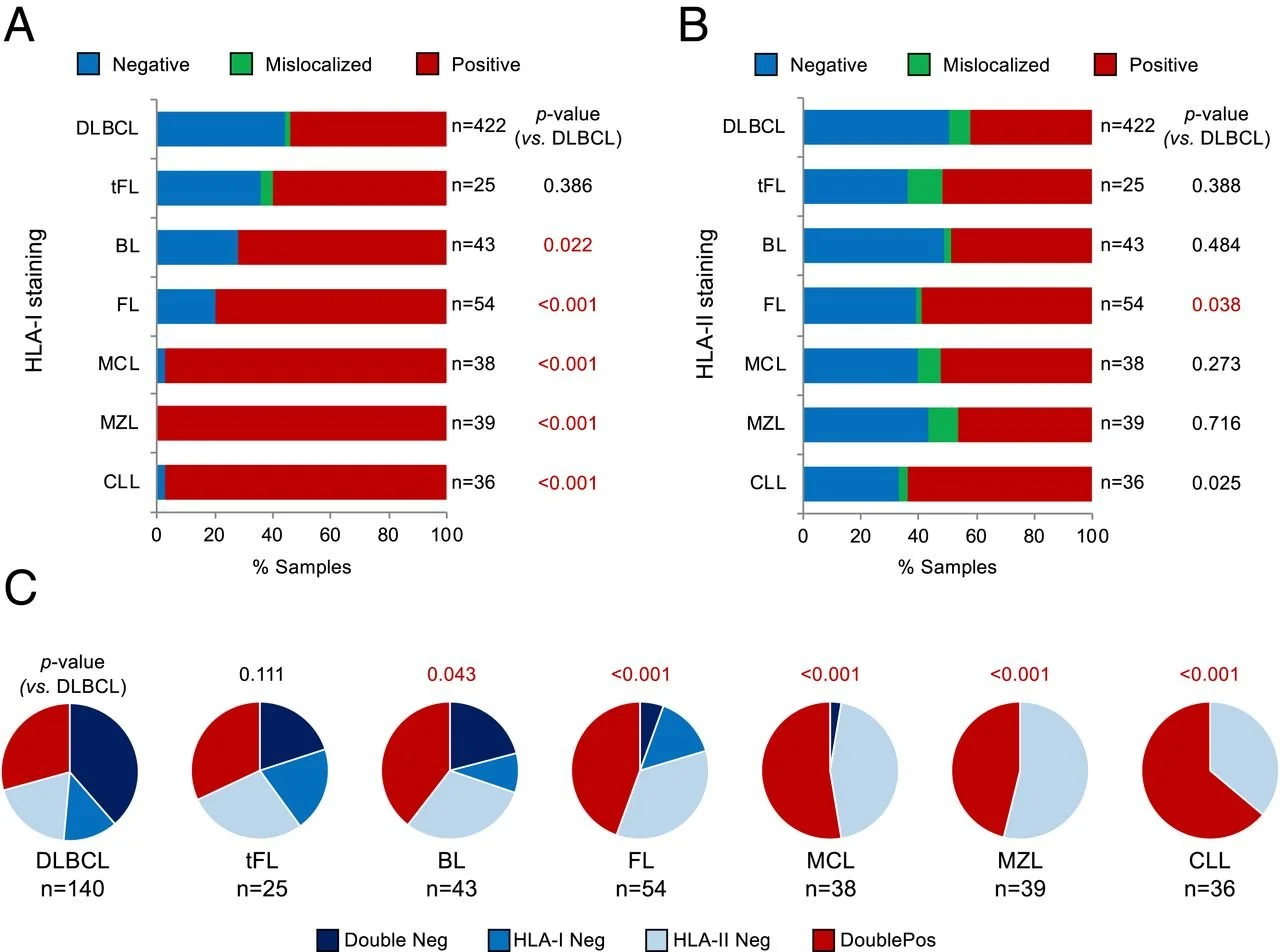
Genetic mechanisms of HLA-I loss and immune escape in diffuse large B cell lymphoma
Fifty percent of diffuse large B cell lymphoma (DLBCL) evade immune-surveillance via somatic genetic lesions abrogating the expression of the class I major histocompatibility complex (MHC-I) complex on the cell surface, thus preventing the presentation of tumor neoantigens to the immune system. The results herein significantly extend these findings by showing that an additional 40% of DLBCL cases, despite expressing MHC-I, carry monoallelic HLA-I genetic alterations that limit the repertoire of neoantigens for presentation to immune cells. Both MHC-I negative and MHC-I positive/monoallelically disrupted cases have significantly higher mutational load. Notably, homozygosis of HLA-I loci is significantly and preferentially enriched in the germline of DLBCL patients, suggesting a stepwise process by which limited neoantigen presentation is selected during DLBCL development.
Single-cell characterization of macrophages in glioblastoma reveals MARCO as a mesenchymal pro-tumor marker
Macrophages are the most common infiltrating immune cells in gliomas and play a wide variety of pro-tumor and anti-tumor roles. However, the different subpopulations of macrophages and their effects on the tumor microenvironment remain poorly understood. We combined new and previously published single-cell RNA-seq data from 98,015 single cells from a total of 66 gliomas to profile 19,331 individual macrophages. Unsupervised clustering revealed a pro-tumor subpopulation of bone marrow-derived macrophages characterized by the scavenger receptor MARCO, which is almost exclusively found in IDH1-wild-type glioblastomas. Previous studies have implicated MARCO as an unfavorable marker in melanoma and non-small cell lung cancer; here, we find that bulk MARCO expression is associated with worse prognosis and mesenchymal subtype. Furthermore, MARCO expression is significantly altered over the course of treatment with anti-PD1 checkpoint inhibitors in a response-dependent manner, which we validate with immunofluorescence imaging. These findings illustrate a novel macrophage subpopulation that drives tumor progression in glioblastomas and suggest potential therapeutic targets to prevent their recruitment.
Pregnancy Specific Glycoproteins: A Possible Mediator of Immune Tolerance of Cancers
Cancer immunotherapy relies upon the immune system recognizing and killing cancer cells. Tumors can elude recognition by readapting existing mechanisms of immune control and suppression. Here we explore the hypothesis that cancers repurpose the immune suppression employed during pregnancy to protect the allogeneic fetus. Those mechanisms are reviewed and shown to be employed both in pregnancy and by tumors. Pregnancy specific glycoproteins (PSGs) produced by fetal trophoblasts are also synthesized by a large number of tumors, which are associated with a poor overall survival of the patient. The family of PSGs may well be a useful target for future checkpoint therapy.
Global Patterns Of Recombination Across Human Viruses
Viral recombination is a major evolutionary mechanism driving adaptation processes, such as the ability of host-switching. Understanding global patterns of recombination could help to identify underlying mechanisms and to evaluate the potential risks of rapid adaptation. Conventional approaches (e.g., those based on linkage disequilibrium) are computationally demanding or even intractable when sequence alignments include hundreds of sequences, common in viral data sets. We present a comprehensive analysis of recombination across 30 genomic alignments from viruses infecting humans. In order to scale the analysis and avoid the computational limitations of conventional approaches, we apply newly developed topological data analysis methods able to infer recombination rates for large data sets. We show that viruses, such as ZEBOV and MARV, consistently displayed low levels of recombination, whereas high levels of recombination were observed in Sarbecoviruses, HBV, HEV, Rhinovirus A, and HIV. We observe that recombination is more common in positive single-stranded RNA viruses than in negatively single-stranded RNA ones. Interestingly, the comparison across multiple viruses suggests an inverse correlation between genome length and recombination rate. Positional analyses of recombination breakpoints along viral genomes, combined with our approach, detected at least 39 nonuniform patterns of recombination (i.e., cold or hotspots) in 18 viral groups. Among these, noteworthy hotspots are found in MERS-CoV and Sarbecoviruses (at spike, Nucleocapsid and ORF8). In summary, we have developed a fast pipeline to measure recombination that, combined with other approaches, has allowed us to find both common and lineage-specific patterns of recombination among viruses with potential relevance in viral adaptation.
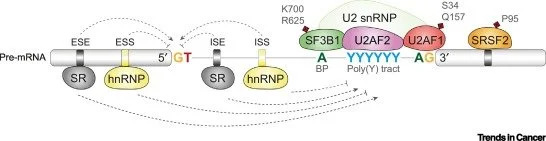
Computing The Role Of Alternative Splicing In Cancer
Accumulating evidence indicates that recurrent spliceosomal mutations contribute to the initiation and progression of several cancers through diverse fundamental cellular processes.A number of computational tools are used to characterize splicing effects in cancer. These tools present limitations that can be overcome by running alternative splicing analysis with multiple tools and integrating the results.Extracting splicing events functionally relevant to cancer requires rigorous quality control to filter technical artifacts, cross-validate the events using independent datasets, and integrate alternative approaches including regulatory network characterization and cancer signaling pathway analyses. By taking advantage of the increasing amount of genomic data, deep learning-based methods have dramatically improved the state-of-the-art performance of alternative splicing analysis.
Mutational and functional genetics mapping of chemotherapy resistance mechanisms in relapsed acute lymphoblastic leukemia
Multiagent combination chemotherapy can be curative in acute lymphoblastic leukemia (ALL). Still, patients with primary refractory disease or with relapsed leukemia have a very poor prognosis. Here we integrate an in-depth dissection of the mutational landscape across diagnostic and relapsed pediatric and adult ALL samples with genome-wide CRISPR screen analysis of gene–drug interactions across seven ALL chemotherapy drugs. By combining these analyses, we uncover diagnostic and relapse-specific mutational mechanisms as well as genetic drivers of chemoresistance. Functionally, our data identify common and drug-specific pathways modulating chemotherapy response and underscore the effect of drug combinations in restricting the selection of resistance-driving genetic lesions. In addition, by identifying actionable targets for the reversal of chemotherapy resistance, these analyses open therapeutic opportunities for the treatment of relapse and refractory disease.
A single-cell atlas of the mouse and human prostate reveals heterogeneity and conservation of epithelial progenitors
Understanding the cellular constituents of the prostate is essential for identifying the cell of origin for prostate adenocarcinoma. Here, we describe a comprehensive single-cell atlas of the adult mouse prostate epithelium, which displays extensive heterogeneity. We observe distal lobe-specific luminal epithelial populations (LumA, LumD, LumL, and LumV), a proximally enriched luminal population (LumP) that is not lobe-specific, and a periurethral population (PrU) that shares both basal and luminal features. Functional analyses suggest that LumP and PrU cells have multipotent progenitor activity in organoid formation and tissue reconstitution assays. Furthermore, we show that mouse distal and proximal luminal cells are most similar to human acinar and ductal populations, that a PrU-like population is conserved between species, and that the mouse lateral prostate is most similar to the human peripheral zone. Our findings elucidate new prostate epithelial progenitors, and help resolve long-standing questions about anatomical relationships between the mouse and human prostate.
Identification of Relevant Genetic Alterations in Cancer using Topological Data Analysis
Large-scale cancer genomic studies enable the systematic identification of mutations that lead to the genesis and progression of tumors, uncovering the underlying molecular mechanisms and potential therapies. While some such mutations are recurrently found in many tumors, many others exist solely within a few samples, precluding detection by conventional recurrence-based statistical approaches. Integrated analysis of somatic mutations and RNA expression data across 12 tumor types reveals that mutations of cancer genes are usually accompanied by substantial changes in expression. We use topological data analysis to leverage this observation and uncover 38 elusive candidate cancer-associated genes, including inactivating mutations of the metalloproteinase ADAMTS12 in lung adenocarcinoma.
A Random Matrix Theory Approach to Denoise Single-Cell Data
Single-cell technologies provide the opportunity to identify new cellular states. However, a major obstacle to the identification of biological signals is noise in single-cell data. In addition, single-cell data are very sparse. We propose a new method based on random matrix theory to analyze and denoise single-cell sequencing data. The method uses the universal distributions predicted by random matrix theory for the eigenvalues and eigenvectors of random covariance/Wishart matrices to distinguish noise from signal. In addition, we explain how sparsity can cause spurious eigenvector localization, falsely identifying meaningful directions in the data. We show that roughly 95% of the information in single-cell data is compatible with the predictions of random matrix theory, about 3% is spurious signal induced by sparsity, and only the last 2% reflects true biological signal. We demonstrate the effectiveness of our approach by comparing with alternative techniques in a variety of examples with marked cell populations.
Genomic Characterization of HIV-Associated Plasmablastic Lymphoma Identifies Pervasive Mutations in the JAK–STAT Pathway
Plasmablastic lymphoma (PBL) is an aggressive B-cell non-Hodgkin lymphoma associated with immunodeficiency in the context of human immunodeficiency virus (HIV) infection or iatrogenic immunosuppression. While a rare disease in general, the incidence is dramatically increased in regions of the world with high HIV prevalence. The molecular pathogenesis of this disease is poorly characterized. Here, we defined the genomic features of PBL in a cohort of 110 patients from South Africa (15 by whole-exome sequencing and 95 by deep targeted sequencing). We identified recurrent mutations in genes of the JAK–STAT signaling pathway, including STAT3 (42%), JAK1 (14%), and SOCS1 (10%), leading to its constitutive activation. Moreover, 24% of cases harbored gain-of-function mutations in RAS family members (NRAS and KRAS). Comparative analysis with other B-cell malignancies uncovered PBL-specific somatic mutations and transcriptional programs. We also found recurrent copy number gains encompassing the CD44 gene (37%), which encodes for a cell surface receptor involved in lymphocyte activation and homing, and was found expressed at high levels in all tested cases, independent of genetic alterations. These findings have implications for the understanding of the pathogenesis of this disease and the development of personalized medicine approaches.
Understanding Coronavirus
Since the identification of the first cases of the coronavirus in December 2019 in Wuhan, China, there has been a significant amount of confusion regarding the origin and spread of the so-called 'coronavirus', officially named SARS-CoV-2, and the cause of the disease COVID-19. Conflicting messages from the media and officials across different countries and organizations, the abundance of disparate sources of information, unfounded conspiracy theories on the origins of the newly emerging virus and the inconsistent public health measures across different countries, have all served to increase the level of anxiety in the population. Where did the virus come from? How is it transmitted? How does it cause disease? Is it like flu? What is a pandemic? What can we do to stop its spread? Written by a leading expert, this concise and accessible introduction provides answers to the most common questions surrounding coronavirus for a general audience.
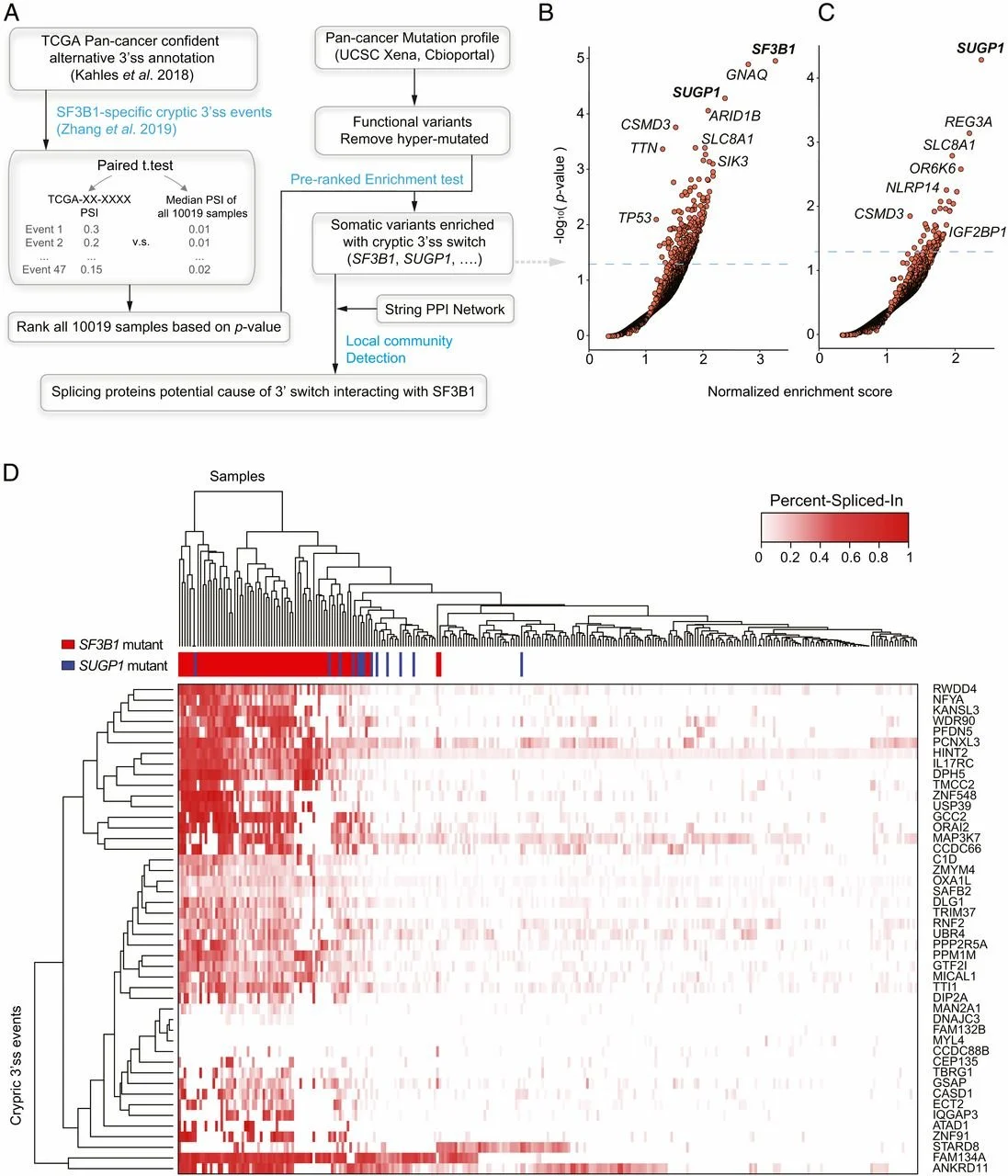
Pan-cancer analysis identifies mutations in SUGP1 that recapitulate mutant SF3B1 splicing dysregulation
The gene encoding the core spliceosomal protein SF3B1 is the most frequently mutated gene encoding a splicing factor in a variety of hematologic malignancies and solid tumors. SF3B1 mutations induce use of cryptic 3′ splice sites (3′ss), and these splicing errors contribute to tumorigenesis. However, it is unclear how widespread this type of cryptic 3′ss usage is in cancers and what is the full spectrum of genetic mutations that cause such missplicing. To address this issue, we performed an unbiased pan-cancer analysis to identify genetic alterations that lead to the same aberrant splicing as observed with SF3B1 mutations. This analysis identified multiple mutations in another spliceosomal gene, SUGP1, that correlated with significant usage of cryptic 3′ss known to be utilized in mutant SF3B1 expressing cells. Remarkably, this is consistent with recent biochemical studies that identified a defective interaction between mutant SF3B1 and SUGP1 as the molecular defect responsible for cryptic 3′ss usage. Experimental validation revealed that five different SUGP1 mutations completely or partially recapitulated the 3′ss defects. Our analysis suggests that SUGP1 mutations in cancers can induce missplicing identical or similar to that observed in mutant SF3B1 cancers.
Mutations in the RNA Splicing Factor SF3B1 Promote Tumorigenesis through MYC Stabilization
Although mutations in the gene encoding the RNA splicing factor SF3B1 are frequent in multiple cancers, their functional effects and therapeutic dependencies are poorly understood. Here, we characterize 98 tumors and 12 isogenic cell lines harboring SF3B1 hotspot mutations, identifying hundreds of cryptic 3′ splice sites common and specific to different cancer types. Regulatory network analysis revealed that the most common SF3B1 mutation activates MYC via effects conserved across human and mouse cells. SF3B1 mutations promote decay of transcripts encoding the protein phosphatase 2A (PP2A) subunit PPP2R5A, increasing MYC S62 and BCL2 S70 phosphorylation which, in turn, promotes MYC protein stability and impair apoptosis, respectively. Genetic PPP2R5A restoration or pharmacologic PP2A activation impaired SF3B1-mutant tumorigenesis, elucidating a therapeutic approach to aberrant splicing by mutant SF3B1.
Here, we identify that mutations in SF3B1, the most commonly mutated splicing factor gene across cancers, alter splicing of a specific subunit of the PP2A serine/threonine phosphatase complex to confer post-translational MYC and BCL2 activation, which is therapeutically intervenable using an FDA-approved drug.
Topological Data Analysis for Genomics and Evolution
Topological Data Analysis for Genomics and Evolution, from Cambridge University Press, explores biology in the age of Big Data. A technical revolution has transformed the field, and extracting meaningful information from large biological data sets is now a central methodological challenge. Algebraic topology is a well-established branch of pure mathematics that studies qualitative descriptors of the shape of geometric objects. It aims to reduce comparisons of shape to a comparison of algebraic invariants, such as numbers, which are typically easier to work with. Topological data analysis is a rapidly developing subfield that leverages the tools of algebraic topology to provide robust multiscale analysis of data sets. This book introduces the central ideas and techniques of topological data analysis and its specific applications to biology, including the evolution of viruses, bacteria and humans, genomics of cancer, and single cell characterization of developmental processes. Bridging two disciplines, the book is for researchers and graduate students in genomics and evolutionary biology as well as mathematicians interested in applied topology.
arcasHLA: high resolution HLA typing from RNAseq
The human leukocyte antigen (HLA) locus plays a critical role in tissue compatibility and regulates the host response to many diseases, including cancers and autoimmune disorders. Recent improvements in the quality and accessibility of next-generation sequencing have made HLA typing from standard short-read data practical. However, this task remains challenging given the high level of polymorphism and homology between HLA genes. HLA typing from RNA sequencing is further complicated by post-transcriptional modifications and bias due to amplification.
To address this, the Rabadan Lab developed arcasHLA, a fast and accurate in silico tool that infers HLA genotypes from RNA sequencing data. Our tool outperforms established tools on the gold-standard benchmark dataset for HLA typing in terms of both accuracy and speed, with an accuracy rate of 100% at two-field resolution for class I genes, and over 99.7% for class II. Furthermore, we evaluate the performance of our tool on a new biological dataset of 447 single-end total RNA samples from nasopharyngeal swabs, and establish the applicability of arcasHLA in metatranscriptome studies.
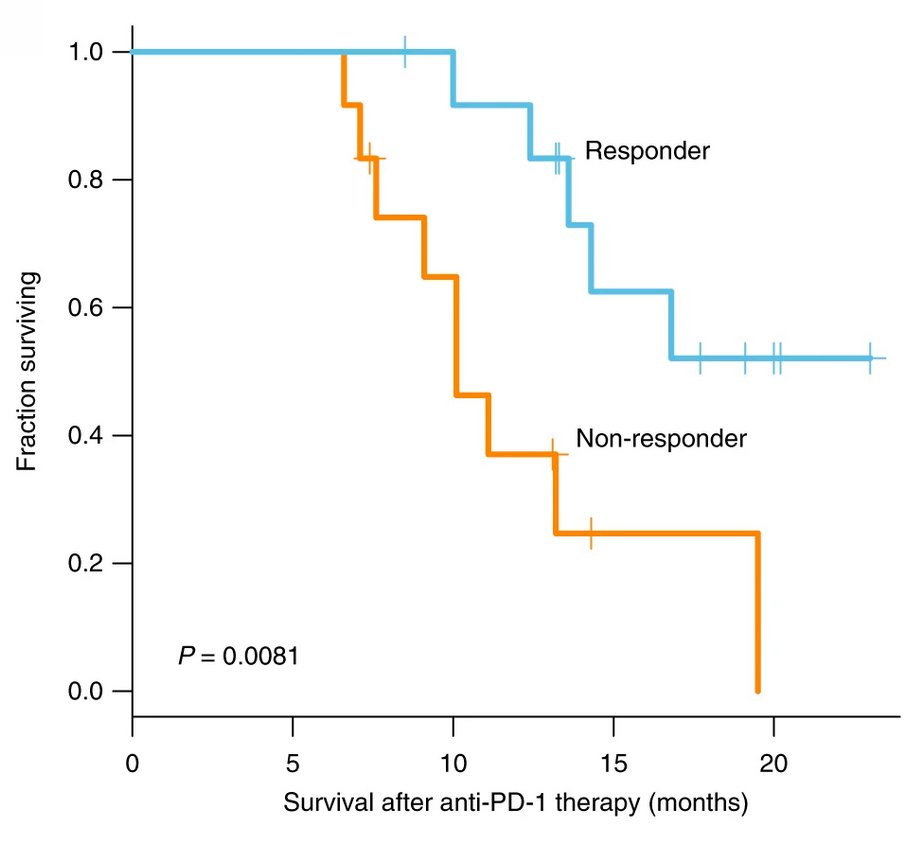
Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma
Immune checkpoint inhibitors have been successful across several tumor types; however, their efficacy has been uncommon and unpredictable in glioblastomas (GBM), where <10% of patients show long-term responses. To understand the molecular determinants of immunotherapeutic response in GBM, we longitudinally profiled 66 patients, including 17 long-term responders, during standard therapy and after treatment with PD-1 inhibitors (nivolumab or pembrolizumab). Genomic and transcriptomic analysis revealed a significant enrichment of PTEN mutations associated with immunosuppressive expression signatures in non-responders, and an enrichment of MAPK pathway alterations (PTPN11, BRAF) in responders. Responsive tumors were also associated with branched patterns of evolution from the elimination of neoepitopes as well as with differences in T cell clonal diversity and tumor microenvironment profiles. Our study shows that clinical response to anti-PD-1 immunotherapy in GBM is associated with specific molecular alterations, immune expression signatures, and immune infiltration that reflect the tumor’s clonal evolution during treatment.
The top figure shows Brain MRIs of two patients treated with nivolumab, one of whom showed disease progression following 2 months of treatment (left, NU 7) while the other showed stable disease without progression after 17 months of treatment (right, NU 11). The bottom figure is a Kaplan–Meier curve comparing overall survival of patients who responded to anti-PD-1 therapy (n = 13) with those that did not respond (n = 12).
Pharmacogenomic landscape of patient-derived tumor cells informs precision oncology therapy
Outcomes of anticancer therapy vary dramatically among patients due to diverse genetic and molecular backgrounds, highlighting extensive intertumoral heterogeneity. The fundamental tenet of precision oncology defines molecular characterization of tumors to guide optimal patient-tailored therapy. Towards this goal, we have established a compilation of pharmacological landscapes of 462 patient-derived tumor cells (PDCs) across 14 cancer types, together with genomic and transcriptomic profiling in 385 of these tumors. Compared with the traditional long-term cultured cancer cell line models, PDCs recapitulate the molecular properties and biology of the diseases more precisely. Here, we provide insights into dynamic pharmacogenomic associations, including molecular determinants that elicit therapeutic resistance to EGFR inhibitors, and the potential repurposing of ibrutinib (currently used in hematological malignancies) for EGFR-specific therapy in gliomas. Lastly, we present a potential implementation of PDC-derived drug sensitivities for the prediction of clinical response to targeted therapeutics using retrospective clinical studies.